-
Notifications
You must be signed in to change notification settings - Fork 154
Separate 1,3 sigmatropic rearrangement family #550
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
8cbc102 to
742f5d6
Compare
|
Tagging @davidfarinajr since I noticed he is working on a PR for 1,3 sigmatropic rearrangement |
 davidfarinajr
left a comment
davidfarinajr
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Thanks for this PR @kspieks ! I've had issues with some of the rates for 1,3 sigmatropic rearrangement family recently, and I think this PR will go a long way towards fixing those issues. I had a wide variety of comments on this review 😂 , some are requested changes (squashing a commit for example) and some are just thinking out loud so don't feel like you need to address them all.
| 2 *3 R!H u0 {1,S} {4,S} | ||
| 3 *1 R!H u0 {1,D} | ||
| 4 *4 R u0 {2,S} | ||
| 4 *4 R!H u0 {2,S} |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
By not allowing *4 to be H, we will no longer generate reactions with *4H-*3C or *4H-*3N (since *3 is only allowed to be O and S for ketoenol family). Do we want to disallow these types of reactions? It seems the barrier is very high for *4H-*3C (~450 kJ/mol for a rxn I calculated here 0f01765) so maybe they will never be relevant and we shouldn't generate them as estimating them to be faster than they are might cause problems
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yea that's a good observation. Technically this PR reduces coverage relative to PR #532. However, the 1,3 sigmatropic template in this PR still expands the coverage of possible reactions that RMG could explore relative to anything before PR #532, so we're not removing legacy reaction templates. Separately, as you point out, these reactions have incredibly high barriers, which almost certainly preclude them from being added to any RMG mechanism. Although we could add training reactions that teach RMG to estimate these ~450 kJ/mol barriers (I had added some in my previous PR), my understanding is that the quality of the estimate (or whether our templates even allow these reactions in the first place) won't have any practical impact since RMG would never include these high barrier reactions.
| @@ -1,8 +1,8 @@ | |||
| #!/usr/bin/env python | |||
| # encoding: utf-8 | |||
|
|
|||
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Yea absolutely :)
| kinetics = ArrheniusBM(A=(3.50759e-62,'s^-1'), n=22.1275, w0=(750402,'J/mol'), E0=(-20429.3,'J/mol'), Tmin=(300,'K'), Tmax=(1500,'K'), uncertainty=RateUncertainty(mu=-0.8800459367924431, var=31.676078103554833, Tref=1000.0, N=56, data_mean=0.0, correlation='Root_N-1R!H-inRing',), comment="""BM rule fitted to 56 training reactions at node Root_N-1R!H-inRing | ||
| Total Standard Deviation in ln(k): 13.494121433083981"""), | ||
| label = "Root_1R!H-inRing", | ||
| kinetics = ArrheniusBM(A=(9.20285e+105,'s^-1'), n=-26.8654, w0=(654062,'J/mol'), E0=(462603,'J/mol'), Tmin=(300,'K'), Tmax=(1500,'K'), uncertainty=RateUncertainty(mu=1.596114397653623, var=262.29968162840566, Tref=1000.0, N=8, data_mean=0.0, correlation='Root_1R!H-inRing',), comment="""BM rule fitted to 8 training reactions at node Root_1R!H-inRing |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
An A of 9e105 and n of -26 concerns me a bit, especially because temperature range is 300-1500K and someone might run RMG hotter than that (I think large negative n is better than large positive when extrapolating to T>1500, but still a bit concerning). Do we know why there is such a big spread here? Which rxns are fit to this node and if there are any outliers?
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
A larger spread is expected higher up the tree, so perhaps we should put constraints on the fitted params (-10<n<10 for example) so we don't get large negative or positive values that might fit better over the T range, but extrapolate more poorly? Or we could expand the T range for fitting, but maybe your training reaction kinetics are not valid over that range, or use the collision limit at high T as the high T rate, not sure.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
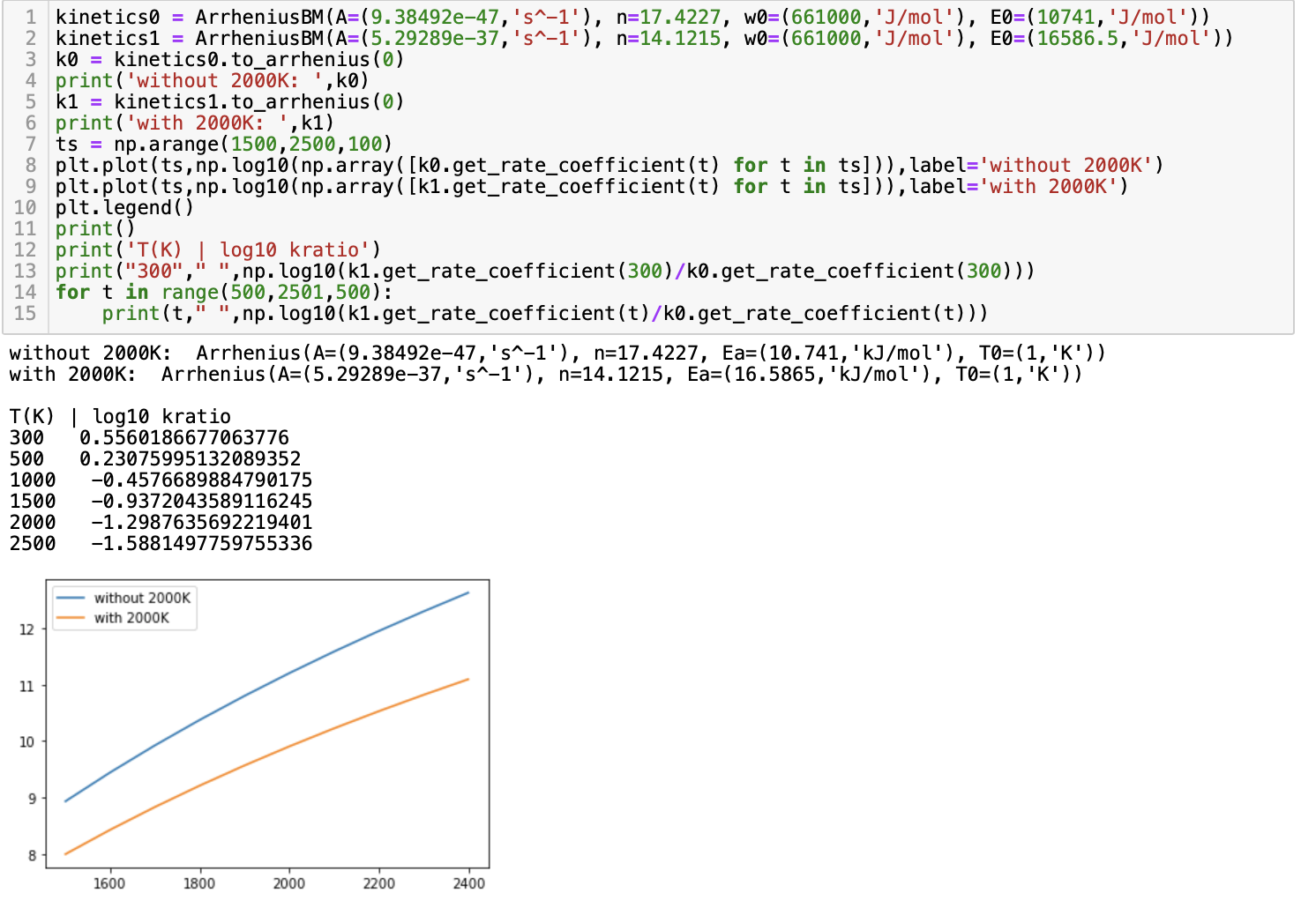
Thanks for bringing this up. To summarize the discussion you, Matt, and I had this morning at RMG developer hours, we ultimately care about the fit quality so it's okay if some of the fitted parameters are odd (very very odd haha) if they give a good fit. Regarding the temperature range, I will override the hardcoded values (link) from the fit_to_reactions method in the ArrheniusBM class to include 2000K. For ketoenol, 14 of the 17 reactions are valid from 300K to 2000K so I'd argue that it makes sense to ask the other 3 training reactions to extrapolate to 2000K rather than restrict all reactions to 1500 K. Similarly, 11 of the 12 training reactions for 1,3 sigmatropic were also fit to 3 parameter Arrhenius from 300K to 2000K. For now, I left these as separate commits so you can see that changes. Including 2000K actually made the fitted parameters more reasonable. I can squash these before you merge
| ['FORM_BOND', '*4', 1, '*1'], | ||
| ]) | ||
|
|
||
| boundaryAtoms = ["*2", "*3"] |
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Do we need boundary atoms? I'm not sure how these are currently used in RMG. Based on the Root, it looks like this would only match a 1,3 rearrangement.
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
Thanks for asking about this at RMG developer hours today. To summarize your discussion, boundary atoms are deprecated. Since ATG, doesn't use them, I will remove this line.
Many reactants in this dataset are not unique. For these cases, only the first instance is included.
|
Thanks for reviewing @davidfarinajr ! I appreciate your comments and have left replies. For now, I pushed fixup commits that I can squash once this is ready to be merged |
742f5d6 to
5d504aa
Compare
|
Thanks for addressing my comments @kspieks ! This PR looks good to me! By including 2000K into the fit, it looks like the rate changes by an order of magnitude over 1500-2000 K (for this particular example anyways), So perhaps we should include 2000K point when fitting BM rules for default Trange settings. I think this is ready to merge, if you want to squash those commits, I'll merge it in. Thanks! |
R!H ensures this family does not overlap with the new ketoenol template Also remove the previous ketoenol training reactions that were previously merged into this template
5d504aa to
694a7dc
Compare
|
Thanks @davidfarinajr! I squashed the commits |
|
I almost forgot about adding |
|
Thanks for reviewing and merging! |
This PR brings several improvements
Improves rate estimates by separating the 1,3 sigmatropic rearrangement reaction template into the traditional ketoenol tautomerization and the 1,3 sigmatropic rearrangement. This essentially reverts PR Rename ketoenol to 1,3 sigmatropic rearrangement (Part 3) #532. The Ea for the simple H transfer with a traditional ketoenol reaction is much lower than the Ea for the heavy atom transfers with 1,3 sigmatropic rearrangement. Combining these trees messed up the rate estimates for ketoenol reactions since higher level nodes were averaging wildly different Arrhenius parameters. Now, the average error for ketoenol rate estimation (last line from running the ATG notebook) is much lower than it was before, and even slightly lower when compared to PR Refit Ketoenol Rate Rules #524 which had not changed the template yet.
Adds a thermo library for all species that had been added as new training reactions for these families. Previously, GAV was used to estimate the thermo for the reverse reaction when fitting the rate tree. The new thermo library will improve the reverse rate estimate when fitting the rate tree. All species thermo was calculated at
CCSD(T)-F12a/cc-pVDZ-F12//ωB97X-D3/def2-TZVPwith AECs and BACs that been added in PRs AEC BAC for ωB97X-D3/def2-TZVP and B97-D3/def2-mSVP #459 and Update CCSD(T)-F12 AECs and BACs #508.Performs a thorough conformer search for all species. No new training reactions were added in this PR. The existing training reactions calculated at
CCSD(T)-F12a/cc-pVDZ-F12//ωB97X-D3/def2-TZVPfrom PRs Refit Ketoenol Rate Rules #524, Refit Ketoenol Rate Rules (Part 2) #527, and Rename ketoenol to 1,3 sigmatropic rearrangement (Part 3) #532 were separated into the new reaction templates. However, a thorough conformer search was done with ACS for all reactants, transition states, and products. The previous PRs that had added these reactions did not do a conformer search because the RDKit functionCalcNumRotatableBondsincorrectly classified these species as rigid. I should've tested it more thoroughly. Upon closer inspection, the documentation's recommended settings do not give the expected result for all molecules, and it is actually better to useRotatableBondSmartsinstead.The zip file contains the Arkane outputs for thermo calculations and Arrhenius fitting. It also contains the notebooks used to fit the rate trees.
notebooks.zip