-
Notifications
You must be signed in to change notification settings - Fork 154
Adding surface libraries #506
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Conversation
|
Thanks for these additions Ting-Chen! I didn't check very carefully but the tests pass and there is a lot here to check by hand. I think one thing we could do on this PR is come up with a new organization scheme for the kinetics libraries surface folder. Because we are adding a bunch of new ones in this PR, it's hard for someone to know which libraries they should use in the Surface folder. Perhaps we could organize it this way? which might looks like Maybe we could add 1 commit here that reorganizes this folder |
|
The Vlachos_Ru0001 library includes the coverage_dependence feature. |
Great! Please put in order a, m, E, and when there is more than one, merge into a single dictionary. Thanks |
|
I reorganized surface libraries to Methane, Ammonia, DOC, and Hydrazine folders. |
|
Run some tests for coverage dependence and the models completed: MODEL GENERATION COMPLETED The final model core has 51 species and 257 reactions MODEL GENERATION COMPLETED The final model core has 16 species and 18 reactions The |
 ChrisBNEU
left a comment
ChrisBNEU
left a comment
There was a problem hiding this comment.
Choose a reason for hiding this comment
The reason will be displayed to describe this comment to others. Learn more.
The reorganizing of the libraries is straightforward and helpful, and the coverage dependent parameters that you added in are all fine. I only skimmed the other reactions, but they have all been used repeatedly so far so I am confident they work. I have also used the schneider pt111 and roldan ir111 libraries for HAN, and it has built the model.
I vote that this is ready to be merged, once we have merged coverage dependence.
24cd544 to
0662f09
Compare

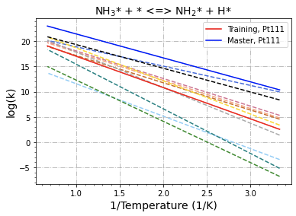
Purpose:
Adding more surface reactions into kinetic libraries
Description:
A separated PR from Add training reactions and enlarge the nodes in the tree for some surface reaction families #479
Separating previous one into three PRs: "adding libraries", "adding tree nodes", and "adding training data"
Notice:
Roldan_Cu111 and Roldan_Ir111 libraries include some bidentate species, need further discussion